SAS Thread - epic egetspec - XMM-Newton
Extraction of spectra in a few clicks: especget
|
Introduction This thread explains how to process spectral data taken with the EPIC cameras using the SAS in a quick, semi-automatic manner. Expected Outcome At the end of this thread a spectrum will have been created together with a background file and compatible detector matrices. These will be in a format where they may be used directly in a spectral fitting package such as XSPEC. SAS Tasks to be Used Prerequisites
Useful Links Last Reviewed: 31 January 2025, for SAS v22.0Last Updated: 30 October 2014 |
Procedure
The SAS task especget has been designed to allow extraction of a source+background and background spectra, a redistribution matrix, and an effective area vector from an EPIC event list with a single command. especget can be launched from the main xmmselect window, in which case an extraction region size optimization algorithm is also ran, which maximizes the signal-to-noise of the net source spectrum. From xmmselect the user is guided by the GUI through the especget steps. This thread illustrates how to use this functionality.
The rebinning options shown in the example above, are by no means the only possible binning available, or the recommended one, more binning options are available in the description of the task specgroup.
- Set up your SAS environment (following the SAS Startup Thread)
- Create an EPIC cleaned and filtered for particle background event file for your observation (see Prerequisites for this thread at the top of the page). Let the name of the filtered file be: EPICclean.fits.
- Start xmmselect,
xmmselect table=EPICclean.fits
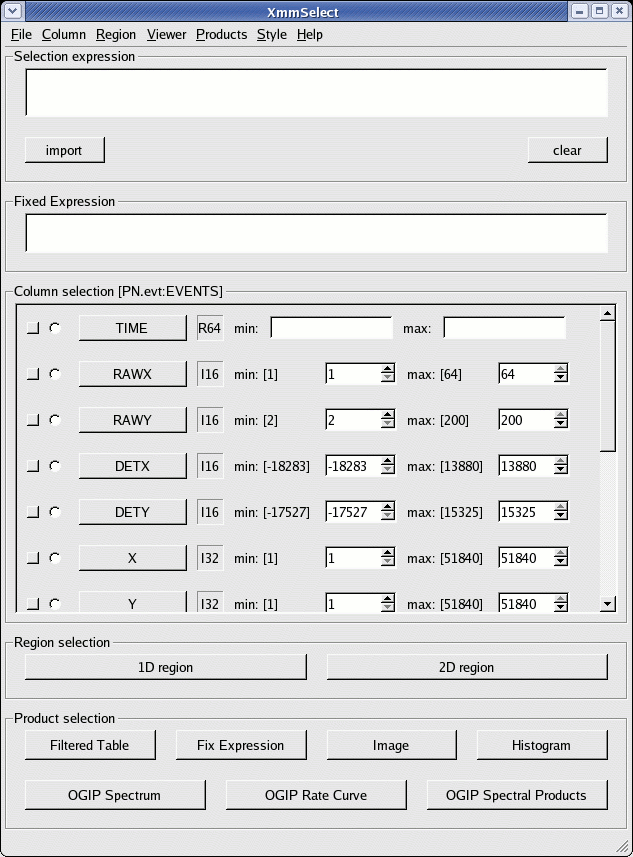
The xmmselect call pops-up a window similar to that shown in Fig.1.
In this window, we identify:
- Extract an image (sky coordinates in this example; extraction in detector - DET[XY] - coordinates is possible as well, and may be preferable for some specific scientific needs). This is accomplished by:
- click the square checkbox besides X and Y in the data column xmmselect panel
- click on Image
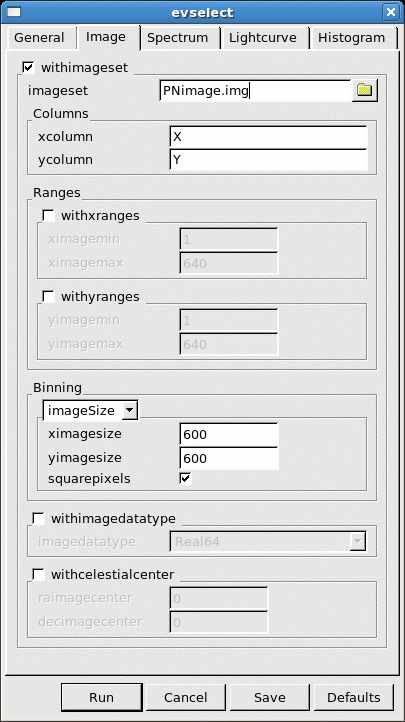
- go the the Image sub-panel in the evselect window (see Fig.2)
- change at least the file name in the imageset window (e.g. to PNimage.fits if we are using an EPIC-pn event file)
- click Run
xmmselect will automatically launch a ds9 window on the created image. - Select the region, from which the source+background spectrum shall be accumulated, using the Region/Circle menu in ds9.
Properties should be set to Source (the green circle on the ds9 window will appear as a solid line) - Select the region, from which the background spectrum shall be accumulated, using the Region/Circle menu in ds9.
Properties should be set to Background (the green circle on the ds9 window will appear as a dashed line) - Click the function button OGIP Spectral Products in the main xmmselect window
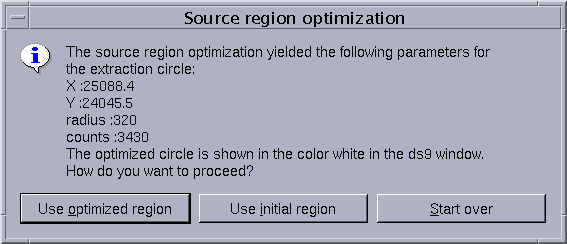
- A window pops-up, asking the user for the preferred method to determine the source+background extraction region:
- Use optimized region: allows the user to extract the source+background spectrum from a circular region, which maximizes the signal-to-noise ratio. The properties of this optimal region are displayed in the window.
- Use initial region: allows the user to extract the source+background spectrum from the region initially chosen.
- Start over: allows the user to start the whole process again, from the definition of spectral extraction regions in the field-of-view image.
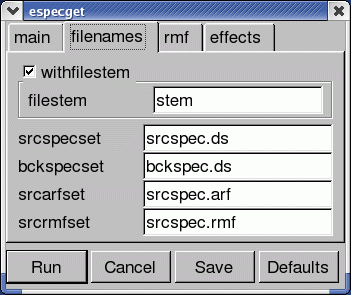
- A current window pops-up, asking for confirmation of the input parameters (see Fig.4). Click Run for producing the spectral products.
The optimum pattern and event flag selections are set automatically for the detector being used (EPIC-MOS or EPIC-pn). If the expression wants to be changed, it can be done through the Main tag of the especget task.
Be reminded that xmmselect silently overwrites files with the same name. The choices in Fig.4 are the defaults. It is recommended to change the filestem parameter to a string of user's choice, or write directly the names of your choice. For example:
- EPICsource_spectrum.fits: source+background spectrum
- EPICbackground_spectrum.fits: background spectrum
- EPIC.rmf: redistribution matrix
- EPIC.arf: effective area vector
- Rebin the spectrum and link associated files. In the following example the spectrum is rebinned in order to have at least 25 counts for each background-subtracted spectral channel and not to oversample the intrinsic energy resolution by a factor larger then 3.
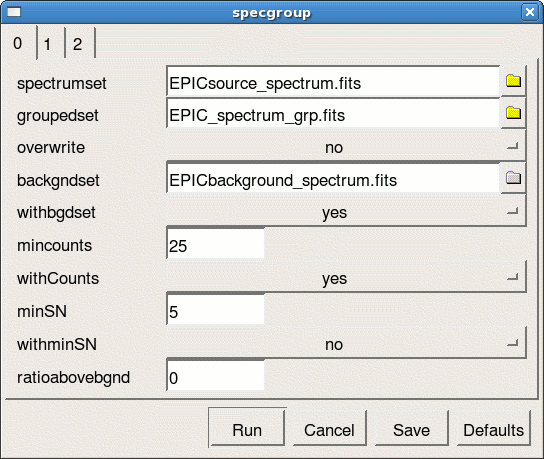
- launch the specgroup GUI interface (see Fig.5)
specgroup -d
- define the following parameters
- spectrumset=EPICsource_spectrum.fits
- groupedset=EPIC_spectrum_grp.fits
- backgndset=yes
- backgndset=EPICbackground_spectrum.fits
- withCounts=yes
- mincounts=25
- withoversampling=yes (on panel 1)
- oversample=3(on panel 1)
- witharfset=yes (on panel 2)
- arfset=EPIC.arf (on panel 2)
- withrmfset=yes (on panel 2)
- rmfset=EPIC.rmf (on panel 2)
- launch the specgroup GUI interface (see Fig.5)
- Everything is now ready to fit the grouped spectrum: EPIC_spectrum_grp.fits.
|
- Removed a total of (1) style text-align:center;
- Removed a total of (11) style text-align:justify;
- Removed a total of (2) border attribute.
- Converted a total of (7) center to div.






































 Sign in
Sign in
 Science & Technology
Science & Technology