SAS Thread - xspec - XMM-Newton
XSPEC Session Thread
Procedure
This thread contains a (very!) simple standard XSPEC session for fitting data from the XMM-Newton X-ray and optical cameras.
EPIC
The EPIC case will be illustrated with an EPIC-pn spectrum (a fully analogous procedure applies to EPIC-MOS spectra as well). We assume in the following that the products have been created and are available in the working directory: a source+background spectrum (PNspectrum.pi), a background only spectrum (PNspectrum_background.pi), a redistribution matrix (PN.rmf), and an ancillary file (PN.arf), both of the later appropriate for the source+background spectrum in use (see Prerequisites for this thread at the top of the page on how to create the spectra and associated files for point-like sources with EPIC-pn or EPIC-MOS). We also assume that the spectrum keywords are not already populated with the response and background files using tasks such as specgroup.
- Start XSPEC
xspec
- Load spectrum data
data 1 PNspectrum.pi
- Load the background spectrum
backgrnd 1 PNspectrum_background.pi
- Load the redistribution matrix
response 1 PN.rmf
- Load the ancillary file
arf 1 PN.arf
- Create the output graphic plotting window
cpd /xs
- Define how data has to be plotted, e.g. in energy bins (wavelength would be a valid alternative)
setplot energy
- Ignore bad channels in data
ignore bad
- Ignore energy intervals where the pn matrices are currently not calibrated (e.g. beyond the energy band 0.2-15.0 keV)
ignore **-0.2 15.-**
(check the EPIC Calibration Status Document for recommended energy ranges to be used for the different modes and EPIC instruments)
- Relax getting a look into your data
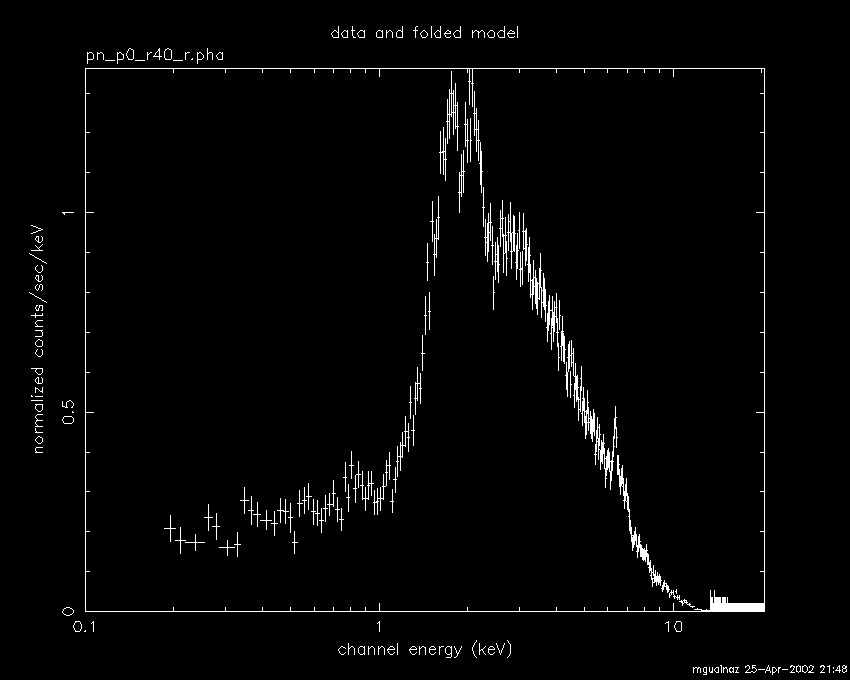
plot data
- Load a model to be fitted (in this case a simple power law model with photoelectric absorption)
model wabs*powerlaw
(you will have to hit return several times for every parameter of the model). The following screen will appear: mo = wa((po)). Model: wabs[1]( powerlaw[2] ) Input parameter value, delta, min, bot, top, and max values for ... Current: 1 0.001 0 0 1E+05 1E+06 wabs:nH> Current: 1 0.01 -3 -2 9 10 powerlaw:PhoIndex> Current: 1 0.01 0 0 1E+24 1E+24 powerlaw:norm> --------------------------------------------------------------------------- --------------------------------------------------------------------------- Model: wabs[1]( powerlaw[2] ) Model Fit Model Component Parameter Unit Value par par comp 1 1 1 wabs nH 10^22 1.000 +/- 0. 2 2 2 powerlaw PhoIndex 1.000 +/- 0. 3 3 2 powerlaw norm 1.000 +/- 0. --------------------------------------------------------------------------- --------------------------------------------------------------------------- Chi-Squared = 2.7158111E+09 using 1675 PHA bins. Reduced chi-squared = 1624289. for 1672 degrees of freedom Null hypothesis probability = 0. - Fit the parameters
fit 100 1e-1
The former number indicates the maximum number of iterations before the minimization routine stops. The latter number indicates the difference in fit statistic between iterations below which the fit is deemed to have converged. - Set the rebinning of your choice (for plotting purposes only!)
setplot rebin 3 4096
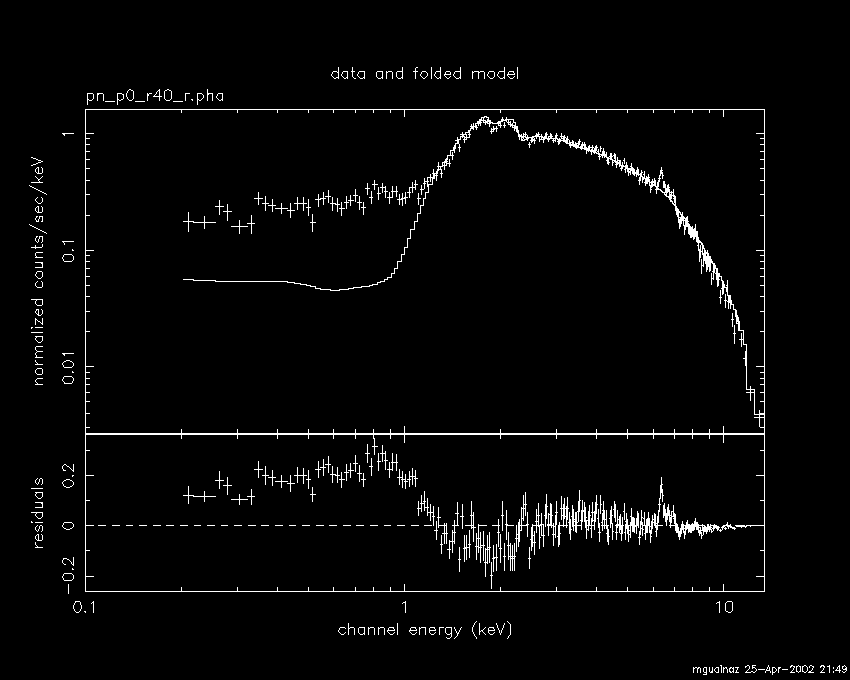
The former number indicates the maximum number of sigmas to be accumulated in a rebinned channel; the latter, the maximum number of channels to be summed. - Plot the data with the model adjustment and e.g. the residuals (alternatives, ratio data/model, absolute chi-squared, etc)
plot data residuals
- Determine one-side errors on the best-fit parameters 1, 2 and 3
error 2.706 1 2 3
The first number indicates the delta chi-squared value.
Notice that the help system in XSPEC is started with help.
RGS
We will assume in the following that products are available in the work directory as produced by SAS rgsproc, the RGS SAS metatask for whole data reduction (see Prerequisites for this thread at the top of the page on How to extract RGS spectra of a point-like source and associated matrices):
- Start XSPEC
xspec
- Load the spectrum data (in this case the RGS1 first order spectrum) into the first internal dataset
data 1:1 /xvsas05/sasval/data/procred/Mkn421/P0099280201R1S001SRSPEC1001.FIT
- Load the corresponding background file:
backgrnd 1 /xvsas05/sasval/data/procred/Mkn421/P0099280201R1S001BGSPEC1001.FIT
- Load the response matrix into the first internal response set. The SAS task rgsrmfgen puts both response and ancillary data into the response matrix file
resp 1 /xvsas05/sasval/data/procred/Mkn421/P0099280201R1S001RSPMAT1001.FIT
- Loading a second dataset into second internal data position (in this case data corresponding to RGS2)
data 2:2 /xvsas05/sasval/data/procred/Mkn421/P0099280201R2S002SRSPEC1001.FIT
- Load the corresponding background file:
backgrnd 2 /xvsas05/sasval/data/procred/Mkn421/P0099280201R2S002BGSPEC1001.FIT
- Load the corresponding response matrix
resp 2 /xvsas05/sasval/data/procred/Mkn421/P0099280201R2S002RSPMAT1001.FIT
A few differences need to be noticed at the data loading level:
- rgsrmfgen (the SAS tasks to generate RGS response matrices, which is run by default in the last stage of rgsproc) creates a response matrix which includes both the redistribution matrix and the effective area vector (ancillary file). There is therefore no need to load any *.arf files in XSPEC when analyzing RGS spectra
- The syntax data n:n allows to keep the fit parameter (fully or partly) independent between the two datasets during the fitting procedure. The alternative syntax data n forces the same model parameters to be fitted to all datasets
- rgsrmfgen (the SAS tasks to generate RGS response matrices, which is run by default in the last stage of rgsproc) creates a response matrix which includes both the redistribution matrix and the effective area vector (ancillary file). There is therefore no need to load any *.arf files in XSPEC when analyzing RGS spectra
- Create the output graphic plotting window
cpd /xs
- Define how the data has to be plotted, e.g. in energy bins (wavelength would be a valid alternative)
setplot energy
- Relax getting a look into your data
plot data
- Ignore bad channels in data
ignore bad
- Ignore energy intervals where the RGS matrices are currently not calibrated (e.g.: beyond 1.9 keV)
ignore 1.9-**
- Load a model to be fitted (in this case a simple power law model)
model powerlaw
(you will have to hit return several times for every parameter of the model)
- Fit the parameters
fit 100 1e-1
The former number indicates the maximum number of iterations before the minimization routine stops. The latter number indicates the difference in fit statistic between iterations below which the fit is deemed to have converged.
- Set the rebinning of your choice (for plotting purposes only!)
setplot rebin 3 4096
The former number indicates the maximum number of sigmas to be accumulated in a rebinned channel. The latter, the maximum number of channels to be summed.
- Plot the data with the model adjustment and e.g the residuals (alternatives, ratio data/model, absolute chi-squared, etc)
plot data residuals
Notice that the help system in XSPEC is started with help.
OM
In order to analyse an OM spectrum with XSPEC, we need: (1) the output spectrum of the task om2pha (see Converting OM data to OGIP II format); (2) the canned response files, one for each filter, that can be downloaded from the XMM-Newton OM response files page. The task om2pha already populated the header keywords of the OM spectrum with the response files names, so there is no need to explicitely insert them in XSPEC. However, in order for the keywords to be read properly by XSPEC, the name of the corresponding canned response files do not have to be changed. Moreover, the files need to be located in the same directory as the OM spectrum. In the following example, we will load simulatenously PN and OM spectra.
- Start XSPEC
xspec
- Load the EPIC-pn spectrum data
data 1:1 PNspectrum.pi
- Load the EPIC-pn background spectrum data
background 1 PNspectrum_background.pi
- Load the EPIC-pn redistribution matrix
response 1 PN.rmf
- Load the EPIC-pn ancillary file
arf 1 PN.arf
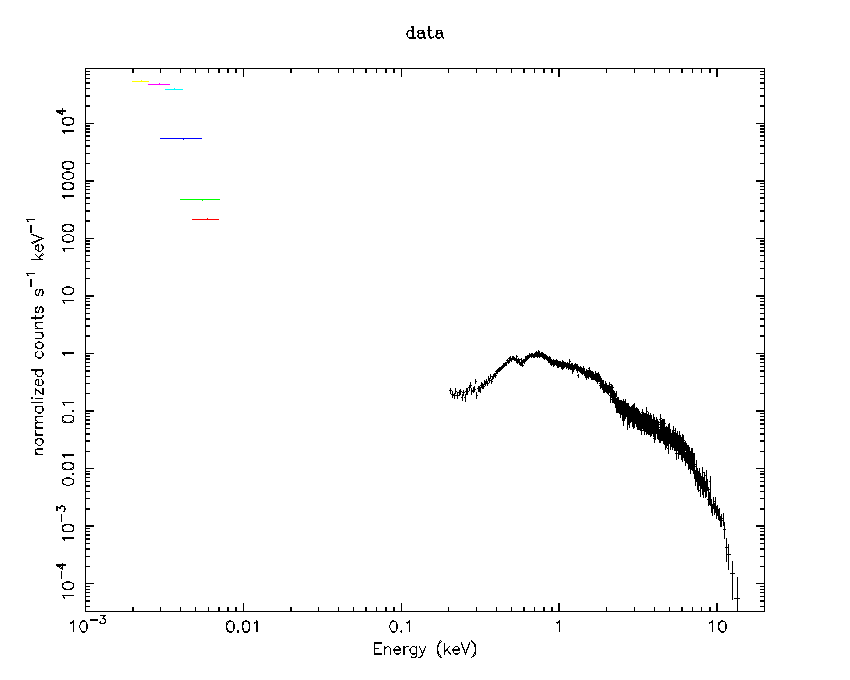
- Load the OM spectrum data (that in this case has six filters)
data 1:2 OMspectrum.pi{1-6}
- Ignore bad channels in data
ignore bad
- Ignore energy intervals where the EPIC-pn matrices are currently not calibrated (e.g. beyond the energy band 0.2-15.0 keV)
ignore 1 **-0.2 15.-**
- Create the output graphic plotting window
cpd /xs
- Define how the data has to be plotted, e.g. in energy bins (wavelength would be a valid alternative)
setplot energy
- Plot the data, e.g. in logarithmic scale
plot ldata
- Proceed loading the model and fitting as described above for EPIC and RGS.
Notice that the help system in XSPEC is started with help.
|
Please, refer to the XSPEC documentation if you wish to really learn XSPEC! If the output products of specgroup are used, the corresponding header keywords are already popullated with the right background and response file names, so all necessary files will be automatically loaded in XSPEC when executing Step 2 above. You can then skip Steps 3-5 and continue in Step 6. |
- Removed a total of (1) style text-align:center;
- Removed a total of (11) style text-align:justify;
- Removed a total of (2) border attribute.
- Converted a total of (5) center to div.






































 Sign in
Sign in
 Science & Technology
Science & Technology